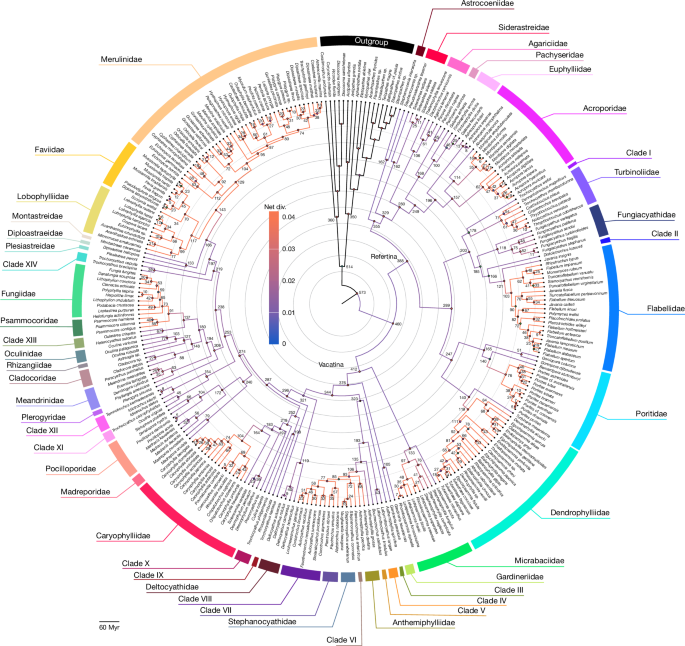
A global coral phylogeny reveals resilience and vulnerability through deep time
Hoegh-Guldberg, O. et al. Coral reefs under rapid climate change and ocean acidification. Science 318, 1737–1742 (2007).
Carpenter, K. E. et al. One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321, 560–563 (2008).
Knowlton, N. Coral reef biodiversity-habitat size matters. Science 292, 1493–1495 (2001).
Pandolfi, J. M., Connolly, S. R., Marshall, D. J. & Cohen, A. L. Projecting coral reef futures under global warming and ocean acidification. Science 333, 418–422 (2011).
Mellin, C. et al. Cumulative risk of future bleaching for the world’s coral reefs. Sci. Adv. 10, eadn9660 (2024).
Kleypas, J. A. et al. Geochemical consequences of increased atmospheric carbon dioxide on coral reefs. Science 284, 118–120 (1999).
Gault, J. A., Bentlage, B., Huang, D. & Kerr, A. M. Lineage-specific variation in the evolutionary stability of coral photosymbiosis. Sci. Adv. 7, eabh4243 (2021).
Stolarski, J. et al. The ancient evolutionary origins of Scleractinia revealed by azooxanthellate corals. BMC Evol. Biol. 11, 316 (2011).
Arrigoni, R. et al. A new sequence data set of SSU rRNA gene for Scleractinia and its phylogenetic and ecological applications. Mol. Ecol. Resour. 17, 1054–1071 (2017).
Quattrini, A. M. et al. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538 (2020).
Kiessling, W. & Simpson, C. On the potential for ocean acidification to be a general cause of ancient reef crises. Glob. Change Biol. 17, 56–67 (2011).
Vasseur, R. et al. Major coral extinctions during the early Toarcian global warming event. Glob. Planet. Change 207, 103647 (2021).
Jacobs, D. K. & Lindberg, D. R. Oxygen and evolutionary patterns in the sea: onshore/offshore trends and recent recruitment of deep-sea faunas. Proc. Natl Acad. Sci. USA 95, 9396–9401 (1998).
Cairns, S. D. Deep-water corals: an overview with special reference to diversity and distribution of deep-water scleractinian corals. Bull. Mar. Sci. 81, 311–322b (2007).
Roberts, J. M., Wheeler, A., Freiwald, A. & Cairns, S. (eds) Cold-Water Corals: The Biology and Geology of Deep-Sea Coral Habitats (Cambridge University Press, 2009).
Orejas, C. et al. Madrepora oculata forms large frameworks in hypoxic waters off Angola (SE Atlantic). Sci. Rep. 11, 15170 (2021).
Campoy, A. N. et al. The origin and correlated evolution of symbiosis and coloniality in scleractinian corals. Front. Mar. Sci. 7, 461 (2020).
McFadden, C. S. et al. Phylogenomics, origin, and diversification of Anthozoans (phylum Cnidaria). Syst. Biol. 70, 635–647 (2021).
Frankowiak, K. et al. Photosymbiosis and the expansion of shallow-water corals. Sci. Adv. 2, e1601122 (2016).
Ries, J. B. Geological and experimental evidence for secular variation in seawater Mg/Ca (calcite-aragonite seas) and its effects on marine biological calcification. Biogeosciences 7, 2795–2849 (2010).
Misof, B. et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767 (2014).
Shao, Y. et al. Phylogenomic analyses provide insights into primate evolution. Science 380, 913–924 (2023).
Zuntini, A. R. et al. Phylogenomics and the rise of the angiosperms. Nature 629, 843–850 (2024).
Erwin, D. H., Valentine, J. W. & Sepkoski, J. J. Jr A comparative study of diversification events: the early Paleozoic versus the Mesozoic. Evolution 41, 1177–1186 (1987).
Scrutton, C. T. The Palaeozoic corals, I: origins and relationships. Proc. York. Geol. Soc. 51, 177–208 (1997).
Scrutton, C. T. & Clarkson, E. N. K. A new scleractinian-like coral from the Ordovician of the Southern Uplands, Scotland. Palaeontology 34, 179–194 (1991).
Ezaki, Y. The Permian coral Numidiaphyllum: new insights into anthozoan phylogeny and Triassic scleractinian origins. Palaeontology 40, 1–14 (1997).
Ezaki, Y. Paleozoic Scleractinia: progenitors or extinct experiments? Paleobiology 24, 227–234 (1998).
Barbeitos, M. S., Romano, S. L. & Lasker, H. R. Repeated loss of coloniality and symbiosis in scleractinian corals. Proc. Natl Acad. Sci. USA 107, 11877–11882 (2010).
Campoy, A. N., Rivadeneira, M. M., Hernández, C. E., Meade, A. & Venditti, C. Deep-sea origin and depth colonization associated with phenotypic innovations in scleractinian corals. Nat. Commun. 14, 7458 (2023).
Lindner, A., Cairns, S. D. & Cunningham, C. W. From offshore to onshore: multiple origins of shallow-water corals from deep-sea ancestors. PLoS ONE 3, e2429 (2008).
Horowitz, J. et al. Bathymetric evolution of black corals through deep time. Proc. R. Soc. B 290, 20231107 (2023).
Rocha, L. A. et al. Mesophotic coral ecosystems are threatened and ecologically distinct from shallow water reefs. Science 361, 281–284 (2018).
Meyer, K. M. & Kump, L. R. Oceanic euxinia in Earth history: causes and consequences. Annu. Rev. Earth Planet. Sci. 36, 251–288 (2008).
Buhl-Mortensen, L., Mortensen, P. B., Armsworthy, S. & Jackson, D. Field observations of Flabellum spp. and laboratory study of the behavior and respiration of Flabellum alabastrum. Bull. Mar. Sci. 81, 543–552 (2007).
Veron, J. E. N. Corals in Space and Time: the Biogeography and Evolution of the Scleractinia (Cornell Univ. Press, 1995).
Ying, H. et al. Comparative genomics reveals the distinct evolutionary trajectories of the robust and complex coral lineages. Genome Biol. 19, 175 (2018).
Stanley, Jr. G. D. & Fautin, D. G. The origins of modern corals. Science 291, 1913–1914 (2001).
Chadwick, N. E. & Adams, C. in Coelenterate Biology: Recent Research on Cnidaria and Ctenophora (eds Williams, R. B. et al.) 263–269 (Springer, 1991).
Daly, M. et al. The phylum Cnidaria: a review of phylogenetic patterns and diversity 300 years after Linnaeus. Zootaxa 1668, 127–182 (2007).
Minter, N. J. et al. Early bursts of diversification defined the faunal colonization of land. Nat. Ecol. Evol. 1, 0175 (2017).
Judd, E. J. et al. A 485-million-year history of Earth’s surface temperature. Science 385, eadk3705 (2024).
Hongzhen, W. & Jianqiang, C. Late Ordovician and early Silurian rugose coral biogeography and world reconstruction of palaeocontinents. Palaeogeogr. Palaeoclimatol. Palaeoecol. 86, 3–21 (1991).
Fedorowski, J. Extinction of Rugosa and Tabulata near the Permian Triassic boundary. Acta Palaeont. Polonica. 34, 47–70 (1989).
Stanley, G. D. Jr. The evolution of modern corals and their early history. Earth Sci. Rev. 60, 195–225 (2003).
Roniewicz, E. & Morycowa, E. Evolution of the Scleractinia in the light of microstructural data. Cour. Forsch. Senckenberg 164, 233–240 (1993).
Anagnostou, E., Huang, K. F., You, C. F., Sikes, E. L. & Sherrell, R. M. Evaluation of boron isotope ratio as a pH proxy in the deep sea coral Desmophyllum dianthus: evidence of physiological pH adjustment. Earth Planet. Sci. Lett. 349, 251–260 (2012).
McCulloch, M. et al. Resilience of cold-water scleractinian corals to ocean acidification: boron isotopic systematics of pH and saturation state up-regulation. Geochim. Cosmochim. Acta 87, 21–34 (2012).
Plusquellec, Y., Webb, G. E. & Hoeksema, B. W. Automobility in Tabulata, Rugosa, and extant scleractinian analogues: stratigraphic and paleogeographic distribution of Paleozoic mobile corals. J. Paleontol. 73, 985–1001 (1999).
Hoeksema, B. W. & Bongaerts, P. Mobility and self-righting by a free-living mushroom coral through pulsed inflation. Mar. Biodivers. 46, 521–524 (2016).
Sentoku, A., Tokuda, Y. & Ezaki, Y. Burrowing hard corals occurring on the sea floor since 80 million years ago. Sci. Rep. 6, 24355 (2016).
Peijnenburg, K. T. et al. The origin and diversification of pteropods precede past perturbations in the Earth’s carbon cycle. Proc. Natl Acad. Sci. USA 117, 25609–25617 (2020).
Kitahara, M. V. Species richness and distribution of azooxanthellate Scleractinia in Brazil. Bull. Mar. Sci. 81, 497–518 (2007).
Capel, K. C. et al. Atlantia, a new genus of Dendrophylliidae (Cnidaria, Anthozoa, Scleractinia) from the eastern Atlantic. PeerJ 8, e8633 (2020).
Kitahara, M. & Cairns, S. Tropical Deep-Sea Benthos Vol. 32 (Publications Scientifiques du Muséum, 2021).
Cairns, S. D. The Marine Fauna of New Zealand: Scleractinia (Cnidaria: Anthozoa) (NIWA, 1995).
Wong, J. S. Y. et al. Comparing patterns of taxonomic, functional and phylogenetic diversity in reef coral communities. Coral Reefs 37, 737–750 (2018).
Seiblitz, I. G. et al. Caryophylliids (Anthozoa, Scleractinia) and mitochondrial gene order: insights from mitochondrial and nuclear phylogenomics. Mol. Phylogenet. Evol. 175, 107565 (2022).
Quattrini, A. M. et al. Universal target-enrichment baits for anthozoan (Cnidaria) phylogenomics: new approaches to longstanding problems. Mol. Ecol. Resour. 18, 281–295 (2018).
Cowman, P. F. et al. An enhanced target-enrichment bait set for Hexacorallia provides phylogenomic resolution of the staghorn corals (Acroporidae) and close relatives. Mol. Phylogenet. Evol. 153, 106944 (2020).
Quek, Z. B. R., Jain, S. S., Neo, M. L., Rouse, G. W. & Huang, D. Transcriptome-based target-enrichment baits for stony corals (Cnidaria: Anthozoa: Scleractinia). Mol. Ecol. Resour. 20, 807–818 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Faircloth, B. C. PHYLUCE is a software package for the analysis of conserved genomic loci. Bioinformatics 32, 786–788 (2016).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000).
Duchêne, D. A., Mather, N., van der Wal, C. & Ho, S. Y. W. Excluding loci with substitution saturation improves inferences from phylogenomic data. Syst. Biol. 71, 676–689 (2021).
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589 (2017).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Anisimova, M., Gil, M., Dufayard, J.-F., Dessimoz, C. & Gascuel, O. Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst. Biol. 60, 685–699 (2011).
Zhang, C., Rabiee, M., Sayyari, E. & Mirarab, S. ASTRAL-III: polynomial time species tree reconstruction from partially resolved gene trees. BMC Bioinform. 19, 15–30 (2018).
Junier, T. & Zdobnov, E. M. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics 26, 1669–1670 (2010).
Mai, U. & Mirarab, S. TreeShrink: fast and accurate detection of outlier long branches in collections of phylogenetic trees. BMC Genom. 19, 23–40 (2018).
Wells, J. W. in Treatise on Invertebrate Paleontology, Part F. Coelenterata (ed. Moore, R. C.) F328–F444 (Geological Society of America, 1956).
Romano, S. L. & Palumbi, S. R. Evolution of scleractinian corals inferred from molecular systematics. Science 271, 640–642 (1996).
Kitahara, M. V. et al. A comprehensive phylogenetic analysis of the Scleractinia (Cnidaria, Anthozoa) based on mitochondrial CO1 sequence data. PLoS ONE 5, e11490 (2010).
Huang, D., Licuanan, W. Y., Baird, A. H. & Fukami, H. Cleaning up the ‘Bigmessidae’: molecular phylogeny of scleractinian corals from Faviidae, Merulinidae, Pectiniidae and Trachyphylliidae. BMC Evol. Biol. 11, 37 (2011).
Stolarski, J. et al. A unique coral biomineralization pattern has resisted 40 million years of major ocean chemistry change. Sci. Rep. 6, 27579 (2016).
Janiszewska, K. et al. Microstructural disparity between basal micrabaciids and other scleractinia: new evidence from Neogene Stephanophyllia. Lethaia 48, 417–428 (2015).
Carbone, F., Matteucci, R., Rosen, B. R. & Russo, A. Recent coral facies of the Indian Ocean coast of Somalia with an interim check list of corals. Facies 30, 1–13 (1994).
Vecsei, A. & Moussavian, E. Paleocene reefs on the Maiella platform margin, Italy: an example of the effects of the Cretaceous/Tertiary boundary events on reefs and carbonate platforms. Facies 36, 123–139 (1997).
Stolarski, J. & Vertino, A. First Mesozoic record of the scleractinian Madrepora from the Maastrichtian siliceous limestones of Poland. Facies 53, 67–78 (2007).
Squires, D. F. The Cretaceous and Tertiary Corals of New Zealand Paleontological Bulletin 29 (New Zealand Geological Survey, 1958).
Bouckaert, R. et al. BEAST 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 15, e1006650 (2019).
Oliveros, C. H. et al. Earth history and the passerine superradiation. Proc. Natl Acad. Sci. USA 116, 7916–7925 (2019).
Sanderson, M. J. Estimating absolute rates of molecular evolution and divergence times: a penalized likelihood approach. Mol. Biol. Evol. 19, 101–109 (2002).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G. & Suchard, M. A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904 (2018).
Brown, J. W. & Smith, S. A. The past sure is tense: on interpreting phylogenetic divergence time estimates. Syst. Biol. 67, 340–353 (2018).
Revell, L. J. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ 12, e16505 (2024).
Höhna, S. et al. RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Syst. Biol. 65, 726–736 (2016).
Tribble, C. M. et al. RevGadgets: an R package for visualizing Bayesian phylogenetic analyses from RevBayes. Methods Ecol. Evol. 13, 314–323 (2022).
Höhna, S. et al. A Bayesian approach for estimating branch-specific speciation and extinction rates. Preprint at BioRxiv https://doi.org/10.1101/555805 (2019).
Vaga, C. F. et al. Data for ‘A global coral phylogeny reveals resilience and vulnerability through deep time’. Figshare https://doi.org/10.6084/m9.figshare.29242487 (2025).
Bosellini, F. R., Papazzoni, C., A. & Vescogni, A. Exceptional development of dissepimental coenosteum in the new Eocene scleractinian coral genus Nancygyra (Ypresian, Monte Postale, NE Italy). Boll. Soc. Paleontol. Ital. 59, 291–298 (2020).
Stolarski, J. On Cretaceous Stephanocyathus (Scleractinia) from the Tatra Mts. Acta Palaeontol. Pol. 35, 31–39 (1990).
First Appeared on
Source link






